|
chr14_-_35873823
|
1.965
|
NM_020529
|
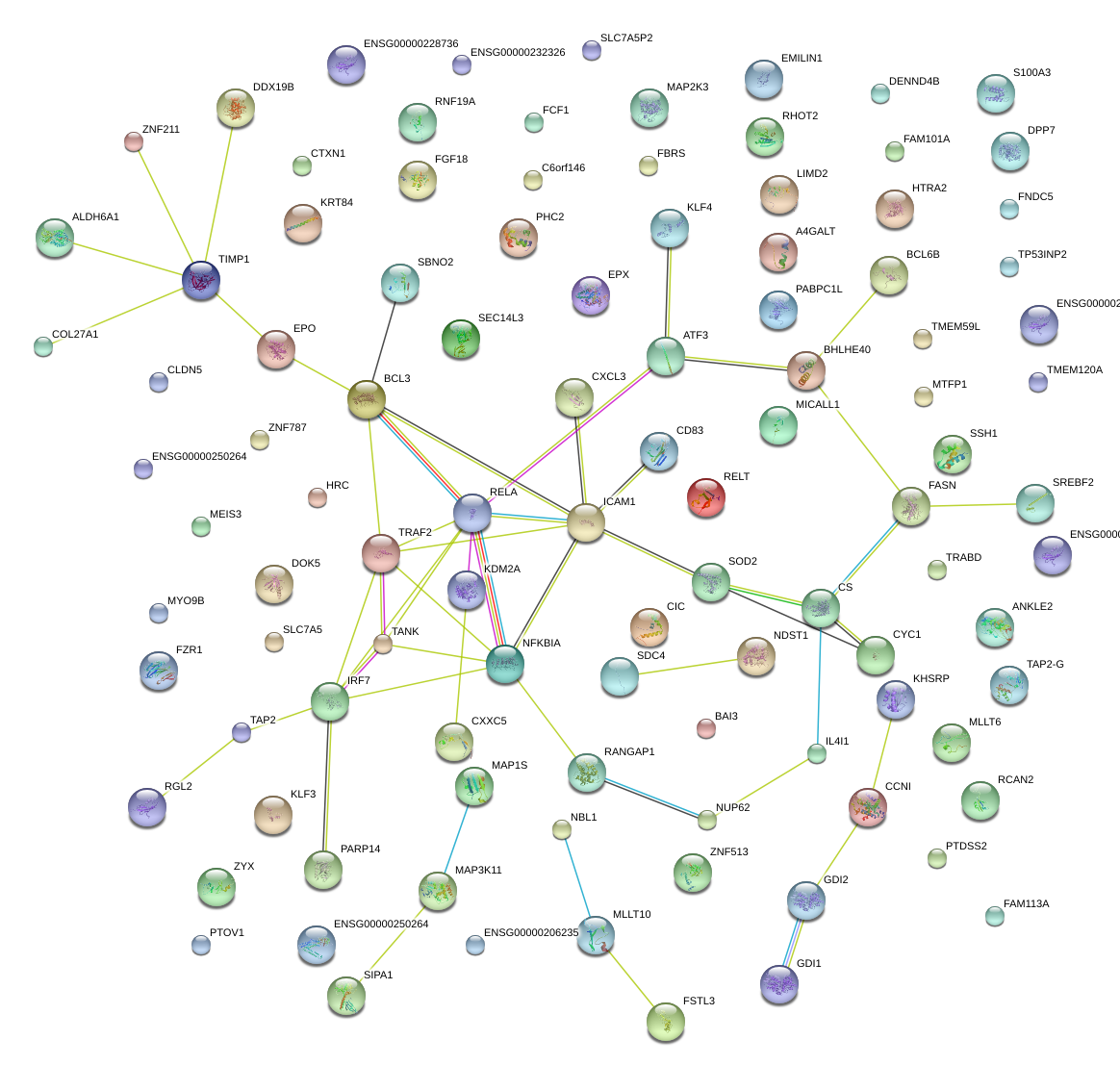
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr11_+_73087670
|
1.412
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr6_+_14118002
|
1.405
|
|
CD83
|
CD83 molecule
|
|
chr19_-_1174201
|
1.359
|
|
|
|
|
chr19_-_1174263
|
1.339
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr2_+_27301693
|
1.184
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr4_-_38666429
|
1.090
|
|
FLJ13197
|
uncharacterized FLJ13197
|
|
chr17_+_36861857
|
1.073
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr12_-_109251333
|
1.054
|
NM_001161330
NM_018984
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr19_-_50432647
|
1.051
|
|
IL4I1
|
interleukin 4 induced 1
|
|
chr19_-_50432672
|
1.049
|
|
IL4I1
|
interleukin 4 induced 1
|
|
chr19_-_1174257
|
1.028
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr7_+_143078399
|
0.999
|
|
ZYX
|
zyxin
|
|
chr19_+_50354417
|
0.998
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_+_10381516
|
0.977
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr19_+_50354368
|
0.976
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_+_17830302
|
0.932
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr19_+_45251933
|
0.923
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr9_-_110251308
|
0.922
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr9_-_110251754
|
0.918
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr22_-_19511777
|
0.890
|
|
CLDN5
|
claudin 5
|
|
chr17_-_61777478
|
0.862
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr19_-_49658645
|
0.856
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr21_+_46825291
|
0.840
|
|
|
|
|
chr11_-_65430214
|
0.838
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr22_-_19512001
|
0.827
|
|
CLDN5
|
claudin 5
|
|
chr3_+_122399665
|
0.824
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr5_+_149865310
|
0.815
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr7_+_143078447
|
0.812
|
|
ZYX
|
zyxin
|
|
chr16_+_30675777
|
0.805
|
NM_001105079
|
FBRS
|
fibrosin
|
|
chr7_+_143078444
|
0.797
|
|
ZYX
|
zyxin
|
|
chr4_-_74904329
|
0.776
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr12_-_52779362
|
0.759
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr11_+_65405538
|
0.750
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr11_+_450346
|
0.748
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr20_+_33292117
|
0.745
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr7_+_143078395
|
0.745
|
|
ZYX
|
zyxin
|
|
chr7_+_143078608
|
0.726
|
|
ZYX
|
zyxin
|
|
chr6_-_32805819
|
0.725
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr22_+_38302296
|
0.714
|
|
MICALL1
|
MICAL-like 1
|
|
chrX_+_153665468
|
0.713
|
|
GDI1
|
GDP dissociation inhibitor 1
|
|
chr11_+_450260
|
0.710
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr2_+_27301434
|
0.707
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr7_-_75623932
|
0.705
|
|
TMEM120A
|
transmembrane protein 120A
|
|
chr2_-_27603584
|
0.697
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr19_-_6424536
|
0.696
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr19_-_7990974
|
0.696
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr19_-_50432679
|
0.691
|
|
NUP62
IL4I1
|
nucleoporin 62kDa
interleukin 4 induced 1
|
|
chr16_-_87903028
|
0.685
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr19_+_676346
|
0.681
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr12_-_109251271
|
0.676
|
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr1_-_153919140
|
0.669
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr20_-_43977023
|
0.666
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr17_+_6926366
|
0.665
|
NM_181844
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr9_-_140009158
|
0.661
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr19_+_17830283
|
0.653
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr19_-_50432734
|
0.645
|
NM_012346
NM_153718
NM_153719
NM_001193357
NM_172374
|
NUP62
IL4I1
|
nucleoporin 62kDa
interleukin 4 induced 1
|
|
chr19_+_17186569
|
0.644
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr22_+_30821610
|
0.642
|
NM_001003704
NM_016498
|
MTFP1
|
mitochondrial fission process 1
|
|
chr12_-_133306531
|
0.636
|
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr11_-_65430427
|
0.627
|
NM_001145138
NM_001243984
NM_001243985
NM_021975
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr6_+_69344939
|
0.623
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr7_+_143078457
|
0.623
|
|
ZYX
|
zyxin
|
|
chr17_-_61777467
|
0.622
|
|
LIMD2
|
LIM domain containing 2
|
|
chr11_-_65381641
|
0.616
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr19_-_56632644
|
0.613
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr11_-_615649
|
0.611
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr1_-_33815498
|
0.610
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr8_+_145149985
|
0.606
|
|
CYC1
|
cytochrome c-1
|
|
chr14_+_93799564
|
0.605
|
NM_020818
|
UNC79
|
unc-79 homolog (C. elegans)
|
|
chr5_+_170846659
|
0.603
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr7_+_143078358
|
0.598
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr17_+_6926414
|
0.597
|
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr3_+_5021096
|
0.593
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr19_+_50353942
|
0.587
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr16_+_718080
|
0.581
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr19_-_50432731
|
0.579
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr9_+_116930778
|
0.579
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr6_-_160114318
|
0.575
|
NM_000636
NM_001024465
NM_001024466
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr8_+_145149936
|
0.574
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr1_-_33336289
|
0.568
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr11_-_615998
|
0.565
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr11_+_66886735
|
0.562
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr19_-_50432678
|
0.559
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr19_+_42788605
|
0.546
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr4_+_38665587
|
0.544
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr7_-_2595327
|
0.538
|
NM_152743
|
BRAT1
|
BRCA1-associated ATM activator 1
|
|
chr6_-_160114186
|
0.534
|
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr7_-_2595112
|
0.531
|
|
BRAT1
|
BRCA1-associated ATM activator 1
|
|
chr6_-_46459010
|
0.524
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_+_5021141
|
0.521
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr9_-_110250707
|
0.516
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr21_+_46293925
|
0.511
|
|
|
|
|
chr6_+_29894105
|
0.508
|
|
|
|
|
chr19_+_18723666
|
0.507
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr8_-_101322180
|
0.504
|
|
RNF19A
|
ring finger protein 19A
|
|
chr17_-_80044975
|
0.497
|
|
FASN
|
fatty acid synthase
|
|
chr2_+_74757243
|
0.496
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_-_47922339
|
0.492
|
|
MEIS3
|
Meis homeobox 3
|
|
chr12_+_124773709
|
0.489
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr9_-_140009169
|
0.484
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr20_+_53092131
|
0.482
|
NM_018431
|
DOK5
|
docking protein 5
|
|
chr22_-_41682187
|
0.480
|
NM_002883
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chr22_-_43116796
|
0.479
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr9_-_140009171
|
0.478
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr6_-_33266297
|
0.477
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr2_+_74757127
|
0.477
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr20_+_43538667
|
0.474
|
NM_001124756
|
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like
|
|
chr22_+_50624353
|
0.473
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr16_+_718157
|
0.471
|
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr22_-_41682169
|
0.469
|
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chr19_+_3506264
|
0.468
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr9_+_139780974
|
0.468
|
|
TRAF2
|
TNF receptor-associated factor 2
|
|
chr22_+_42229137
|
0.467
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr5_+_139060295
|
0.466
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_+_212781969
|
0.462
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_74756986
|
0.460
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_-_50432952
|
0.458
|
NM_016553
|
NUP62
|
nucleoporin 62kDa
|
|
chr17_+_21188021
|
0.457
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr9_-_140009162
|
0.457
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr9_-_110252045
|
0.452
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_+_10381763
|
0.449
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr6_-_4079390
|
0.449
|
NM_173563
|
C6orf146
|
chromosome 6 open reading frame 146
|
|
chr6_+_31588523
|
0.448
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr1_+_19972259
|
0.447
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr20_-_2821245
|
0.447
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr1_-_153521717
|
0.443
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr3_+_5021305
|
0.441
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr7_+_100318422
|
0.438
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr13_+_37006628
|
0.435
|
|
CCNA1
|
cyclin A1
|
|
chr6_+_31588444
|
0.434
|
NM_004638
NM_080686
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr17_-_79829174
|
0.433
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr19_+_10362886
|
0.433
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr5_+_131593388
|
0.432
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr21_+_46825001
|
0.432
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr6_+_14117857
|
0.430
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr11_+_67806517
|
0.427
|
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr13_+_37006408
|
0.423
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr19_+_39390567
|
0.423
|
NM_002503
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr11_-_2017822
|
0.423
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_+_212782103
|
0.422
|
|
ATF3
|
activating transcription factor 3
|
|
chr17_-_79829272
|
0.421
|
NM_001185078
NM_004309
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr19_+_10362912
|
0.418
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr17_-_27467395
|
0.417
|
|
MYO18A
|
myosin XVIIIA
|
|
chr17_-_2614926
|
0.416
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr19_-_6424820
|
0.415
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr17_+_36861398
|
0.415
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr7_-_75623961
|
0.413
|
NM_031925
|
TMEM120A
|
transmembrane protein 120A
|
|
chr19_+_10362598
|
0.411
|
NM_015956
NM_146387
NM_146388
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr20_-_10654139
|
0.409
|
|
JAG1
|
jagged 1
|
|
chr17_-_48207113
|
0.409
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr3_-_48672807
|
0.409
|
|
SLC26A6
|
solute carrier family 26, member 6
|
|
chr6_+_31588524
|
0.406
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr18_+_57567191
|
0.406
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr11_-_59436488
|
0.405
|
NM_152716
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr7_-_37488554
|
0.404
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_+_7792034
|
0.403
|
NM_001005273
NM_005852
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr18_-_77711603
|
0.403
|
|
PQLC1
|
PQ loop repeat containing 1
|
|
chr15_+_52311438
|
0.402
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr11_+_45825873
|
0.401
|
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr19_+_7587436
|
0.399
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr21_+_40177847
|
0.398
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr11_-_2018446
|
0.397
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr15_+_52311432
|
0.396
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr17_+_38137185
|
0.396
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr9_-_130661886
|
0.396
|
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr19_+_39390570
|
0.394
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr11_-_71159371
|
0.392
|
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr6_+_29855506
|
0.391
|
|
|
|
|
chr16_-_4838316
|
0.391
|
NM_001154458
NM_144605
|
SEPT12
|
septin 12
|
|
chr11_-_71159442
|
0.391
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr6_+_29910246
|
0.390
|
NM_001242758
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr5_-_139944045
|
0.389
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr19_+_13906207
|
0.389
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr17_-_79829237
|
0.387
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr20_-_2821120
|
0.386
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr6_+_32936400
|
0.386
|
NM_005104
|
BRD2
|
bromodomain containing 2
|
|
chr15_+_52311408
|
0.386
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr2_-_27603249
|
0.383
|
NM_001201459
|
ZNF513
|
zinc finger protein 513
|
|
chr16_+_67694795
|
0.380
|
NM_001037281
NM_016948
|
PARD6A
|
par-6 partitioning defective 6 homolog alpha (C. elegans)
|
|
chr20_+_32399046
|
0.379
|
NM_176812
|
CHMP4B
|
charged multivesicular body protein 4B
|
|
chr11_-_57092283
|
0.379
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr7_+_155089614
|
0.379
|
|
INSIG1
|
insulin induced gene 1
|
|
chr17_-_79828691
|
0.377
|
NM_001185077
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr11_-_1785104
|
0.374
|
|
CTSD
|
cathepsin D
|
|
chr5_-_180237114
|
0.373
|
NM_001114617
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr20_+_45523523
|
0.371
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr1_-_44820923
|
0.362
|
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr12_+_102271349
|
0.361
|
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr9_+_139780939
|
0.360
|
NM_021138
|
TRAF2
|
TNF receptor-associated factor 2
|
|
chr7_+_44646220
|
0.360
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr19_+_39899749
|
0.359
|
|
|
|
|
chr6_+_126111782
|
0.359
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_+_74757116
|
0.358
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_-_47249629
|
0.357
|
|
STRN4
|
striatin, calmodulin binding protein 4
|
|
chr17_+_21188032
|
0.356
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr20_+_61904158
|
0.355
|
NM_018209
NM_175609
|
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1
|
|
chr12_+_102271082
|
0.355
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr19_+_50432399
|
0.355
|
NM_001193646
|
ATF5
|
activating transcription factor 5
|
|
chr22_+_42229105
|
0.354
|
NM_004599
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|